A new system, which consists of a large LLM and a network of agentic tools, outperformed several other models and human physicians [1].
Too rare to easily diagnose
Rare diseases can be notoriously hard to diagnose. Patients average over 5 years to receive a correct diagnosis, enduring repeated referrals, misdiagnoses, and unnecessary interventions in what is known in rare disease medicine as ‘the diagnostic odyssey’ [2]. These rare diseases, defined as conditions affecting fewer than 1 in 2,000 people, collectively impact over 300 million people worldwide. About 7,000 distinct disorders of this type have been identified, with 80% of them being genetic in origin [3].
While AI assistants have shown great promise in diagnostics, diagnosing rare diseases remains a daunting task even for them. Rare diseases are often multisystemic and require cross-disciplinary knowledge; individual diseases have very few cases, making supervised learning hard; and hundreds of new rare genetic diseases are discovered per year, so knowledge is constantly shifting. On top of that, clinical deployment of such models demands transparent reasoning rather than black-box predictions.
In a new study published in Nature, an international team of scientists has presented DeepRare, a multi-agent system for differential diagnosis of rare diseases. While based on the large language model DeepSeek-V3, the system is different from a basic LLM in that it integrates more than 40 specialized agentic tools for various tasks.
Not your usual LLM
DeepRare uses a three-tier design. Tier 1 is the Central Host, a large LLM with a memory bank. It orchestrates the entire workflow: decomposes the diagnostic task, decides which agents to invoke, synthesizes evidence, makes tentative diagnoses, and runs self-reflection loops. Tier 2 is the Agent Servers layer. It consists of six specialized modules, each managing its own tools, such as the Phenotype Extractor, which converts free-text clinical narratives into standardized terms, and the Knowledge Searcher, which retrieves data in real time from web search engines and medical-specific sources. Retrieved documents are then summarized and relevance-filtered by a lightweight LLM (GPT-4o-mini). The external data sources the agents use, such as Google, PubMed, and Wikipedia, constitute Tier 3.
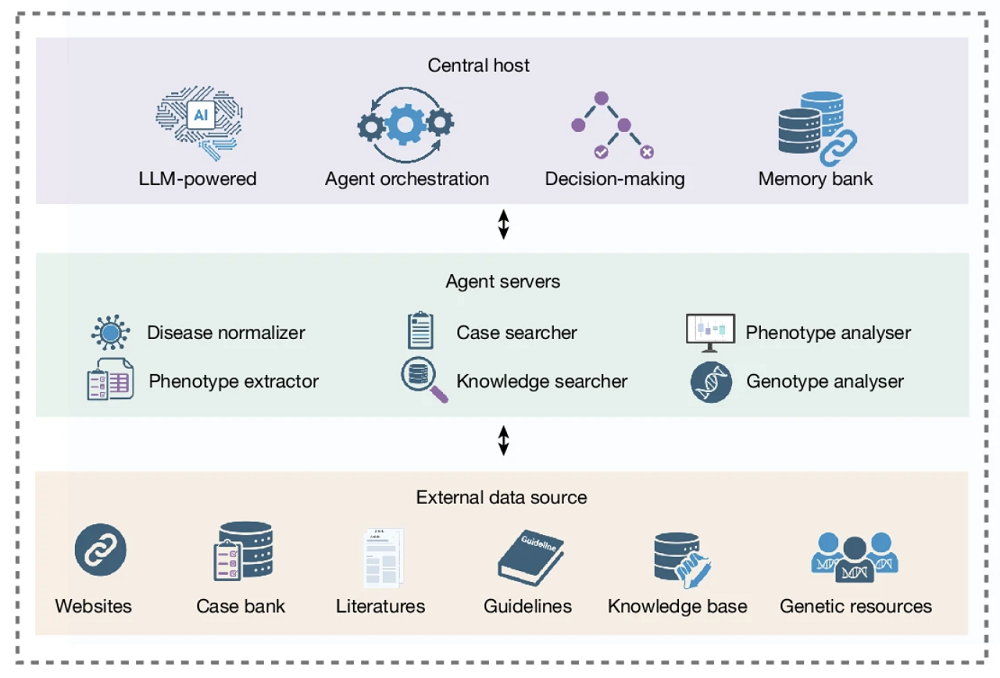
The system operates in two stages. The first one is information collection, where phenotype and genotype branches run in parallel. The phenotype branch standardizes HPO (Human Phenotype Ontology) terms, retrieves relevant literature and cases, and runs phenotype analysis tools. The genotype branch annotates variants and ranks them by clinical significance. The central host then performs synthetic analysis and generates a tentative diagnosis list.
The second stage is self-reflective, where the central host critically re-evaluates each hypothesis against all collected evidence. If all candidates are ruled out during self-reflection, the system goes back, increases the search depth, collects more evidence, and repeats as needed. Once candidates survive self-reflection, the system generates a final ranked list of diseases with reasoning chains (free-text rationales with clickable reference links).
DeepRare’s crucial advantage is that it does not have to be pre-trained on cases of rare diseases, as training LLMs requires a lot of data which simply does not exist for rare diseases, some of which are only known from a handful of cases. Instead, a generally trained LLM orchestrates specialized tools for data retrieval and analysis, synthesizes their outputs through reasoning, and iteratively validates its own conclusions.
Best in class
DeepRare was evaluated across nine datasets, spanning 6,401 total cases, 2,919 distinct rare diseases, and 14 medical specialties. The metrics used were Recall@1, @3, and @5 (whether the correct diagnosis appears in the top 1, 3, or 5 predictions).
The first evaluation was against 15 other digital tools, including general LLMs, reasoning LLMs, medical LLMs, and agentic systems. All the models received standardized Human Phenotype Ontology (HPO) descriptions as input.
DeepRare achieved an average Recall@1 of 57.18% and Recall@3 of 65.25% across all benchmarks, outperforming the second-best method (Claude-3.7-Sonnet-thinking) by 23.79% and 18.65% margins, respectively. However, given the pace of LLM development, several models released after the study’s design period were not included in the comparison; for instance, the top ChatGPT version the study used was 4o.
The model then went head-to-head with human expert physicians. 163 clinical cases were presented identically to DeepRare and five rare disease physicians with at least 10 years of experience, who were allowed to use search engines but not AI tools. DeepRare achieved Recall@1 of 64.4% vs. physicians’ 54.6% and Recall@5 of 78.5% vs. 65.6%. According to the authors, this is one of the first demonstrations of a computational model surpassing expert physicians on rare disease phenotype-based diagnosis.
To validate DeepRare’s reasoning, the researchers then turned to ten associate chief physicians, who evaluated 180 randomly sampled cases. They assessed whether each cited reference was both reliable and directly relevant to the diagnostic conclusion and found reference accuracy to be 95.4%.
Literature
[1] Zhao, W., Wu, C., Fan, Y., Qiu, P., Zhang, X., Sun, Y., Zhou, X., Zhang, S., Peng, Y., Wang, Y., Sun, X., Zhang, Y., Yu, Y., Sun, K., & Xie, W. (2026). An agentic system for rare disease diagnosis with traceable reasoning. Nature, 10.1038/s41586-025-10097-9. Advance online publication.
[2] Glaubitz, R., Heinrich, L., Tesch, F., Seifert, M., Reber, K. C., Marschall, U., … & Müller, G. (2025). The cost of the diagnostic odyssey of patients with suspected rare diseases. Orphanet Journal of Rare Diseases, 20(1), 222.
[3] Nguengang Wakap, S., Lambert, D. M., Olry, A., Rodwell, C., Gueydan, C., Lanneau, V., … & Rath, A. (2020). Estimating cumulative point prevalence of rare diseases: analysis of the Orphanet database. European journal of human genetics, 28(2), 165-173.
View the article at lifespan.io








































